Introduction
Two guidelines were recently published to refine the diagnostic criteria of acute myeloid leukemia (AML): the 5th edition of the World Health Organization Classification of Haematolymphoid Tumors (WHO-HAEM5, PMID: 35732831) and the International Consensus Classification (ICC; PMID: 35767897), and both expanded molecularly defined AML subtypes. In this study, we utilized a cohort of 1135 AML cases to examine their performance.
Methods
From Sep 1, 2018 to Dec 31, 2022, a consecutive cohort of 1135 cases diagnosed with de novo AML according to WHO-HAEM4R (PMID: 27069254) were enrolled, with a median follow-up of 20 months. All underwent high-throughput sequencing (PMID: 34135310, 29932212).
Results
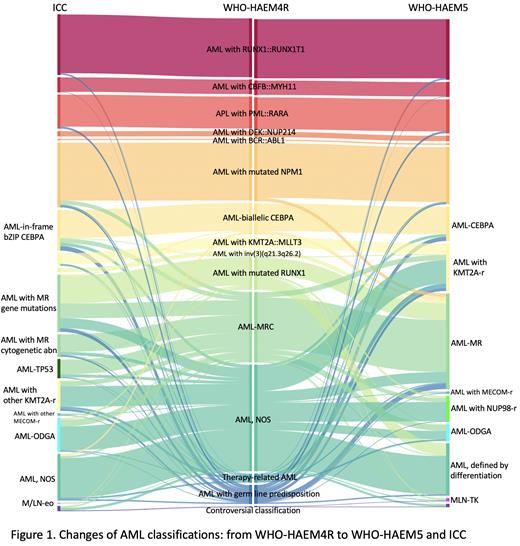
When classified according to WHO-HAEM4R, 55% of the 1135 cases were diagnosed as AML with recurrent genetic abnormalities, 16% were AML with myelodysplasia-related changes (AML-MRC), 2% were therapy-related AML, 4% were AML with germline predisposition, and 24% fell into AML not otherwise specified (AML, NOS).
Major changes in WHO-HAEM5 include the inclusion of AML subtypes of KMT2A rearrangement (KMT2A-r), MECOM-r, and NUP98-r; adding AML with other defined genetic alterations (AML-ODGA) as a basket entity; and annulling the entity of AML with mutated RUNX1 (AML-RUNX1). The entity of AML with CEBPA mutation (AML-CEBPA) has changed to include not only biallelic (biCEBPA) but also single bZIP (smbZIP-CEBPA) mutations. AML-MRC in WHO-HAEM4R is redefined as AML, myelodysplasia-related (AML-MR), with a mutation-based definition based on an 8-gene panel (without RUNX1).
When comparing WHO-HAEM4R and WHO-HAEM5 (Fig. 1), AML with KMT2A-r increased from 2% to 10% because WHO-HAEM5 no longer restricts the partner genes of KMT2A. AML with NUP98-r and AML-ODGA in WHO-HAEM5 account for 6% and 4%, respectively. AML-MR increased from 12% AML-MRC to 21% benefited from the 8-gene panel-based definition. AML-RUNX1 has been annulled in WHO-HAEM5 and 63% cases of this subtype were reclassified as AML-MR, and 30 cases with RUNX1 mutations as the only basis for classification were reclassified as AML defined by differentiation. In total, AML without defining genetic abnormalities dropped from 24% to 11%.
When comparing WHO-HAEM4R and ICC (Fig. 1), AML with mutated TP53 (AML-TP53) is a new category in ICC and accounted for 4% of AML, 79% of them were previously classified as AML-MRC in WHO-HAEM4R. AML-MR in WHO-HAEM5 is divided into two entities in ICC: AML with MR gene mutations (including RUNX1) and AML with MR cytogenetic abnormalities, comprising 13% and 5% of cases, respectively. ICC only includes in-frame bZIP CEBPA mutations. Thus 6 cases classified as AML-biCEBPA based on WHO-HAEM4R fell into AML, NOS under ICC since their CEBPA mutations were not in bZIP. Conversely, 21 cases with smbZIP-CEBPA and diagnosed as AML, NOS according to WHO-HAEM4R were classified as AML-CEBPA under ICC.
Cases classified as AML-CEBPA based on WHO-HAEM5 tended to have a better overall survival (OS) than cases classified as AML-biCEBPA under WHO-HAEM4R because AML-smbZIP-CEBPA was added while cases with MR gene mutations which had a worse prognosis fell into AML-MR under WHO-HAEM5. Despite ICC only included in-frame bZIP-CEBPA mutations, the prognosis of AML-CEBPA diagnosed under ICC was not superior to that under WHO-HAEM5. This might be because cases with both MR cytogenetic abnormalities and CEBPA mutations were reclassified as AML-CEBPA under ICC's hierarchical principle. ICC distinguished AML with KMT2A::MLLT3 from AML with KMT2A-r, while WHO-HAEM5 only included AML with KMT2A-r entity. Survival analysis showed significantly better OS in AML with KMT2A::MLLT3 than AML with KMT2A-r.
ICC separated AML-TP53 from AML-MR due to the notorious prognosis of TP53 defects, and cases classified as AML-TP53 showed the worst OS in this cohort. But prognostic analysis showed no difference between AML-TP53 and cases with concurrent TP53 mutation and other AML-defining genetic abnormalities. The rationality of considering AML-TP53 as a distinct entity remains debatable.
Conclusions
Both new classification systems consider genomic features more extensively and enhance the precise classification of AML. Discrepancies between WHO-HAEM5 and ICC, like classifying AML with RUNX1 mutations and the rationality of defining AML-TP53 as a unique entity, remain open questions that require further research for clearer answers.
Disclosures
No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal